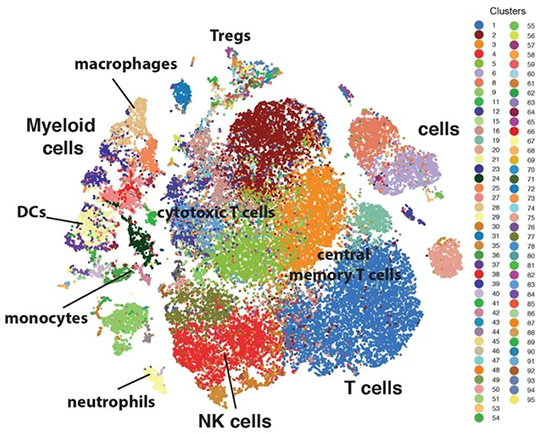
Traditional genome and transcriptome sequencing technologies are typically performed on a sample cell population, masking the differences between individual cells. Single-cell transcriptome sequencing is a new technology for high-throughput RNA sequencing at the single-cell level. It can obtain the transcriptome expression data at the single-cell level, understand the cell heterogeneity information of the sample, realize cell population division and difference analysis, cell evolution process and cell interaction analysis, and depict high-resolution cell maps.
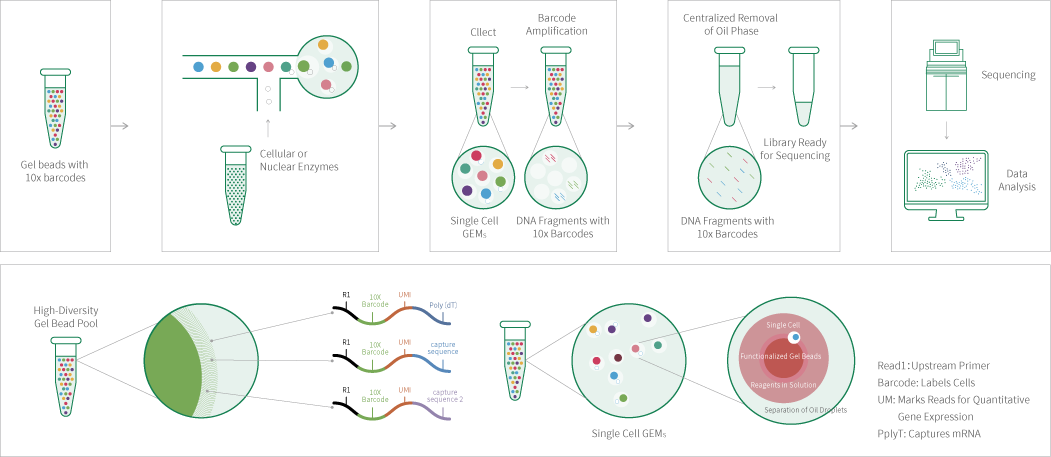
Utilizing the principle of droplet-based methods, individual cells are sorted through microfluidics and mixed with gel beads carrying barcodes and UMIs (Unique Molecular Identifiers), enzymes, and partitioning oil to generate single-cell emulsion droplets. Each droplet serves as an independent reaction system. Within the droplets, the gel beads dissolve and the cells lyses to release mRNA, which by reverse transcription produces cDNA with barcode and unique molecular tag for sequencing. Following the disruption of the oil layer surrounding the droplets, the cDNA is amplified, and a cDNA library is prepared. This library is subsequently sequenced using an Illumina sequencing platform, allowing for the acquisition of gene expression profiles from a large number of individual cells.
Cell Throughput:
Up to 8 samples can be simultaneously detected in a single experiment
Each sample can capture up to tens of thousands of cells.
Short Detection Time:
Preparation of cell suspensions, single-cell capture, amplification, and library construction can all be completed within one day.
High Cell Capture Efficiency:
Single cell capture efficiency up to 65%.
High Cell Compatibility:
No restrictions on cell size or type.
Nuclei of cells with diameters exceeding 40μm can also be detected.
Simple and Convenient:
Integrates single-cell sorting, amplification, and library construction into one process, featuring high automation and requiring no manual operation.
10x Genomics' single-cell sequencing technology is primarily applied in the research of single-cell gene expression profiles, cellular heterogeneity, and functional differences. It can also be used to study single-cell gene expression profiles of cells at different developmental stages and under various disease conditions.
Identification of novel cell types
Construction of cell atlas
Biomarker research
Disease classification studies
Tumor cell heterogeneity and immunemzmicroenvironment research
Mechanisms of disease onset and progression
Immune system development and differentiation
